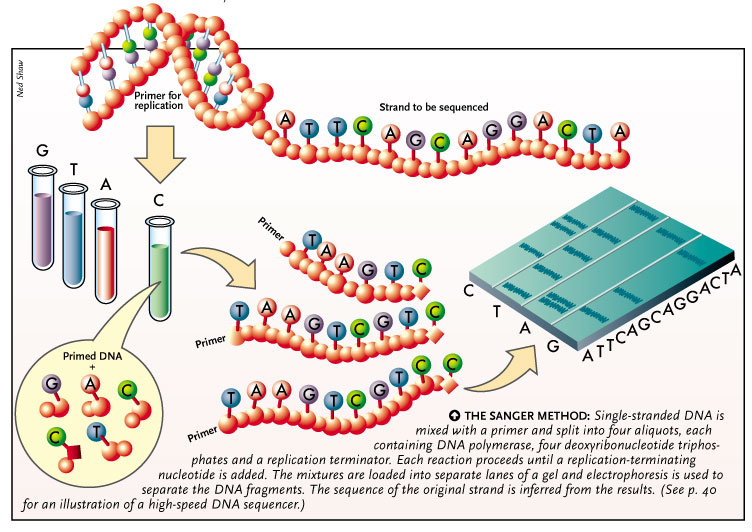
Single-stranded DNA is mixed with a primer and split into four aliquots, each containing DNA polymerase, four deoxyribonucleotide triphos-phates and a replication terminator. Each reaction proceeds until a replication-terminating nucleotide is added. The mixtures are loaded into separate lanes of a gel and electrophoresis is used to for an illustration of a high-speed DNA sequencer.)
What's happening in the DNA sequencing industry? After all, the human genome sequence is done and dusted. Two big players in the industry have recently undergone major changes: Michael W. Hunkapillar, who developed the technology that made high-throughput sequencing possible, stepped down in August from his role as president of Applied Biosystems. And Amersham was purchased by General Electric this year and rebranded as GE Healthcare. Upstart companies are developing a new wave of technologies to challenge capillary technology, currently the only game in town. For the time being it is business...
WHERE ARE WE NOW?
The human genome stole most of the headlines, but scientists around the world have been busily sequencing genomes of hundreds of species over the last decade. And they continue to do so.
In August, the National Human Genome Research Institute (NHGRI) announced the latest batch of initiates to the sequencing hall of fame, a list that includes nine mammals and nine nonmammals. Some of the mammals, including the African savannah elephant, the domestic cat, and the orangutan, provide an injection of charisma; others, such as the European common shrew and the rabbit, are less alluring. But all play an important part in helping researchers to interpret the human genome. (
Likewise, the nine nonmammalian organisms will help to elaborate the evolution of anatomy, physiology, development, and behavior. This role of honor goes to a slime mold, a ciliate, a choanoflagellate, a placozoan, a cnidarian, a snail, two roundworms, and the lamprey.
The ongoing, massive sequencing effort requires hundreds of high-speed DNA sequencers, each costing in the neighborhood of $300,000 (US). J. Craig Venter's company, Celera Genomics, one of the two groups that produced a draft human genome, used at least 300 sequencers to do the job. Applied Biosystems, which has an estimated 70% of the sequencer market, installed 10,000 automated sequencers in the past decade, according to Research and Markets, a Dublin-based firm. (Applera Corporation now comprises the Celera Genomics group and the Applied Biosystems group.)
The need for new sequencing capacity may be maxed out, however. "The drive to actually set up new high-throughput genome centers to carry out high-throughput sequencing is not there to the same degree as it was when the drive was on to sequence the human genome," says Andy Bertera, vice president of core products at GE Healthcare, the second largest supplier of DNA sequencers.
"There was a big drive and a lot of investment to sequence the human genome. Then, once we sequenced the human genome, public interest sort of waned a little bit. However, there's still a lot of sequencing to be done," says Bertera. "There's a new animal or new plant or some new organism that is sequenced almost on a weekly to monthly basis," he adds.
The J. Craig Venter Science Foundation Joint Technology Center is one of the world's largest DNA sequencing facilities. It has 100 Applied Biosystems 3730xl sequencers and has no current plans to buy more, despite its ability to accommodate as many as 400, says Heather Kowalski, vice president of policy and planning at the Center for the Advancement of Genomics in Rockville, Md. "Our current average output is approximately two billion bases per month versus the 0.09 billion bases per month in 1999," Kowalski says. "We are also testing several new sequencing technologies at the facility in the hopes that eventually something will replace or at least significantly augment the output of the traditional capillary array sequencer."
So, while Applied Biosystems reported a 6% increase in revenues in the fourth quarter of 2004, to $460.5 million, sales of its DNA sequencing products declined. Large genome centers aren't buying, but that doesn't make sequencers a dead-end market. "Outside of the genome centers, we see that demand is strong," says Philippe Nore, Applied Biosystems senior director of strategic planning for genetic analysis products. "People are doing more and more sequencing, finding more and more adaptations with our sequencers, such as genotyping or resequencing."
Resequencing certain parts of the genome to detect single nucleotide polymorphisms (SNPs) helps researchers understand specific genes or diseases and has become a core function for sequencing systems. Although microarrays can also be used to resequence, DNA sequencers have a much larger installed base, according to Applied Biosystems, which markets both types of instruments. "Microarray platforms are limited to resequencing only, whereas sequencers can perform de novo sequencing and SNP genotyping as well," a company spokesperson says.
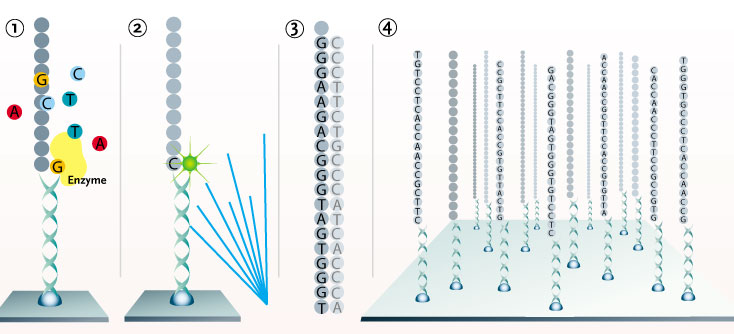
Solexa; redrawn by Ned Shaw
1. DNA is processed into single-stranded fragments and attached to a single molecule array with anchors and primers. Fluorescently labeled nucleotides pair with the first base of each fragment and are added to the primer by a polymerase. The remaining free nucleotides are removed. 2. Laser light causes the nucleotides to fluoresce, and a CCD camera scans the entire array to identify the incorporated nucleotides on each fragment. The fluorescence is then removed. 3. This cycle of incorporation, detection, and identification is repeated about 25 times to determine the first 25 bases in each fragment. 4. By simultaneously sequencing all molecules in the array, the first 25 bases for the hundreds of millions of fragments are determined. These sequences are aligned and compared to determine single nucleotide polymorphisms (SNPs) and other genetic variations.
Bertera points out that the market is interested in seeing how researchers will use sequence information. "Interpretation is critical. Resequencing of parts of the human genome to understand specific diseases [associated with] SNPs is obviously important," Bertera says. "That tends to take you in the direction of diagnostics, and diagnostics sequencing is really in its infancy."
NEXT STOP, THE CLINIC?
DNA sequencing needs to be easier and cheaper before it is clinic-ready. Currently, sequencing an entire mammalian genome costs from $10 to $50 million, but the hope is that a wave of new sequencing technologies will bring the cost down to $100,000, and possibly even to $1,000. The National Institutes of Health called for applications in February for grants to develop new, low-cost sequencing technologies for just this purpose. Capillary electrophoresis systems reign supreme in accuracy, although they tend to be relatively slow and expensive and can read only a few hundred bases per reaction.
A number of promising technologies are being explored, including the single-molecule array. DNA fragments are anchored to the surface of an array, fluorescent-labeled nucleotides are added, and then the array is scanned. The technique could potentially cut costs and speed up DNA sequencing, because it requires little or no sample preparation and uses fewer expensive reagents. But, the technology is still being developed.
One of the firms working on a single-molecule array approach is Essex, UK-based Solexa. "The approach drastically reduces, and at best obviates, the need for complicated and costly sample preparation with the consequential reduction in laboratory preparation and reagent overheads," says Solexa's business development director Simon Bennett. He says the firm's systems would ultimately be capable of four to five orders of magnitude improvement in efficiency, cost, and throughput over conventional sequencing technology (see Table on p. 47).
Helicos Biosciences in Cambridge, Mass., also is working on a single-molecule sequencing approach. The firm licensed its core technology from Stephen Quake, then a researcher at California Institute of Technology and now at Stanford University. "The power of the method is the high degree of parallelism. We can do hundreds of millions of strands at a time," says Stan Lapidus, president and CEO of Helicos.
GenoVoxx of Lübeck, Germany, is working on a single-molecule method called AnyGene, in which DNA fragments deposited on a two-dimensional array are subjected to sequencing-by-synthesis.
One firm that is not pursuing the single-molecule approach is Branford, Conn.-based 454 Life Sciences, a subsidiary of New Haven, Conn.-based CuraGen. 454 Life Sciences' technology uses beads that are attached to DNA strands, which are placed in individual wells of its PicoTiterPlate. PCR occurs in each well, and the sequence of each DNA strand is determined by a sequencing-by-synthesis method. Based on this technique, the company obtained a $2.4 million grant from the NHGRI.
Experts anticipate that DNA sequencing of whole genomes for clinical purposes using these new technologies will likely occur in the next couple of decades. Their use in patient care, however, will initially be restricted. "Between 10 and 20 years from now, it will be increasingly common for patients who are diagnosed with specific diseases or are suspected of having specific diseases to have a full DNA fingerprint taken," Lapidus says. But the goal of sequencing healthy individuals to identify disease risks is a longer way off. He adds, "I think it will not be common yet to simply do, as a matter of course, a complete base end-to-end readout on somebody who is asymptomatic for any given disease."
IMPROVING CAPILLARY ELECTROPHORESIS
While these technologies offer a glimpse at the future of DNA sequencing, some industry observers question how soon these products will deliver on their promises. "I'm sure in the future some of these technologies will [make sequencing] super-cheap," says Jason Kramer, manager of the DNA Sequencing Core at the Dana-Farber/Harvard Cancer Center in Boston. "But, I don't see [some of the sequencing facilities] changing for quite some time," he says.
The companies that pioneered commercial DNA sequencing also have their sights set on the future. The Applied Biosystems group is working on new applications for its existing capillary electrophoresis-based instruments, with a particular interest in pharmacogenomics. It also is working on making the instruments more economical for DNA sequencing.
"We don't think we're done yet with the [current] technology. There is still a lot of improvement we can bring ... [and] ways to lower the overall cost," says Nore. Applied Biosystems plans to lower the cost of sequencing with its systems by making sample preparation more efficient. According to the firm, sample prep accounts for roughly 50% of the cost of sequencing.
Bertera says GE Healthcare has been working with its customers at genome centers in identifying ways to make DNA sequencing more efficient and cost-effective. "We are updating, for example, our MegaBACE 4000 instrument, which is the only 384-capillary instrument on the market, to develop an instrument called the MegaBACE 4500." This will allow customers to upgrade their existing instrumentation and give them longer read-lengths, Bertera adds. He notes that the company also has a certified-preowned program aimed at lowering the costs for researchers who want to do DNA sequencing, but who might not have the financial resources to purchase new instruments. Such instruments come from labs that are moving from sequencing into functional biology or proteomics.
So, the DNA sequencers that brought you the human genome aren't likely to gather dust anytime soon. Cheaper sequencing is eagerly awaited as newer technologies are pitted against improvements in the current standards.
Edward Winnick
NEED A GENOME SEQUENCED? THINK OUTSOURCING
So you want an organism sequenced, but you don't happen to have a $300,000 sequencer handy, or a core facility at your disposal. Who do you call? A wide variety of companies will be more than happy to help you out, for a price. Among the largest for-profit companies that conduct genomic sequencing are Agencourt Bioscience of Beverly, Mass., and SeqWright and Lark Technologies, both in Houston. A few dozen companies do this work, and most have been around for about a decade.
"It takes a big investment up front for all the equipment, personnel, training, and overhead [to sequence an organism]," says Ken Paynter, operations manager at SeqWright, a molecular biology research support firm. "A lot of researchers just aren't doing enough volume of sequencing themselves where it's cost effective for them to maintain their own department," he adds. Because of the market pressures to reduce costs, which subsidized universities and government labs often don't encounter, such companies become highly efficient at sequencing.
"This is what we do, and we're experts at it," says Paul McEwan, vice president and co-chief scientific officer for Agen-court, which provides genomic services. "We have a tremendous amount of capacity," he says, "so often we can turn these projects around much more quickly than someone can do themselves."
Paynter and McEwan estimate that full-length sequencing of a bacterium might require anywhere from six months to a few years to complete, while a draft sequencing that covers most (though not all) of a genome might take a few months. As for cost, "if you're talking about an entire bacterium, it's typically a six-figure type of project," McEwan says. Paynter agrees that large-scale sequencing can run "a couple of million" dollars. Prices tend to drop the bigger a project is, because the cost per sample decreases.
Some turn to outside companies because their own sequencing facilities are backed up with more complicated work. Victor Velculescu, assistant professor of oncology at the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University, turned to Agencourt to help with his cancer research, which requires sequence-intensive analysis. Although Johns Hopkins has its own sequencing facility, it tends to be used for more complicated and time-consuming projects. "For this kind of application, where it's not so crucial that each sample be done right, but we simply need quantity, it's an optimal thing to outsource because the chances of somebody messing things up are small," Velculescu says. "And because our current pipeline is full, we really have no choice but to source out."
Others aren't sold on the idea of out-sourcing genomic sequencing. "There is still much research and development that remains to be done in sequencing, and there are parts of it that remain an art," says Aristides Patrinos, associate director of the Department of Energy's (DOE) Office of Biological and Environmental Research in Germantown, Md. "It's not at the stage where you can do this like you do more routine things."
The DOE's Production Genomics Facility in Walnut Creek, Calif., recently began taking on projects selected through a peer-review process. Says Patrinos: "We believe the scientist who has a good idea and is given a grant – but his progress depends on having a specific sequence – should be able to get that sequence free of charge."
- Dana Wilkie
FOCUS ON FEW TECHNOLOGIES
All of these technologies aim to cut down on the cost and increase the efficiency of DNA sequencing through massively parallel high-throughput sequencing. The major drawback to all of these new technologies is short read-lengths. Traditional Sanger sequencing utilizing capillary electrophoresis is still considered the most accurate and reliable method for DNA sequencing and is likely to retain that distinction in the near-term.
Solexa
Location: Essex, UK.
Founded: Spun out of the University of Cambridge in 1998.
Technology: Single-molecule sequencing utilizing DNA cluster technology. Arrays will be capable of sequencing 100 million sample DNA templates per cm2 of chip.
Read Length: 25–30 bases.
Expected Launch: Expects to be sequencing genomes next year.
Helicos BioSciences
Location: Cambridge, Mass.
Founded: May 2003.
Technology: Single-molecule sequencing in which up to 300 million fragments can be attached to a single slide, enabling 10x coverage of the human genome in a single experiment.
Read Length: 5–10 bases now, but working on increasing that to 25 bases.
Expected Launch: Undisclosed.
GenoVoxx
Location: Lübeck, Germany.
Founded: Spun out of the University of Lübeck in May 2002.
Technology: AnyGene technology utilizing the sequencing-by-synthesis method.
Read Length: 45 nucleotides, but with a 30% error rate.
Expected Launch: Aiming for 2006.
454 Life Sciences
Location: Branford, Conn.
Founded: Spun out of Curagen in 2000.
Technology: Sequencing-by-synthesis method whereby beads are attached to individual strands of DNA and placed in individual wells of its PicoTiterPlate.
Read Length: Short read-lengths. Company researchers have achieved production read-lengths of 100 bases.
Expected Launch: Q1 2005.
SEQUENCING: WHERE IS THE FUNDING?
Four years ago, the National Human Genome Research Institute (NHGRI) used 83% of its $150 million sequencing budget on mapping the human genome. In 2003, 27% of its $163 million budget was devoted to the human genome, and in 2004, less than 1%. "Now we are using the sequencing budget on dozens of other organisms, which will tell us more about the structure and function of humans," says NHGRI spokesman Geoff Spencer.
Now that the human genome has been sequenced, federal funding has steadily shifted toward the larger and more complex task of sequencing the genomic DNA of Earth's multiple other life forms. The government's new focus includes microbes, bacteria, and fungi, as well as more complex plants and animals.
In 2000, more than 90% of the $50 million devoted to sequencing at the US Department of Energy went toward human DNA. This year, the DOE, which has its own "Genomes to Life" program, is spending $54 million, almost all of which is devoted to nonhuman genome sequencing. Aristides Patrinos, director of the DOE's Office of Biological and Environmental Research in German-town, Md., says his office's "focus has shifted mostly to microbial genomes and other model organisms we have interest in."
Genomics research in some form is supported in several directorates at the National Science Foundation. The 2004 budget includes $12 million for the"Assembling the Tree of Life" program, the foundation's long-term effort to map the relationships among living things by using DNA comparisons of different organisms. The project aims to build a family tree to help scientists better understand how life evolved on Earth.
The NSF began a push for plant genome sequencing in 1998 with $40 million for funding individual research projects and centers. This year, funding for the Plant Genome Research Program is nearly $90 million, while the NSF's Microbial Genome Sequencing Project is funded with $15 million, and the Arabidopsis 2010 Project garnered $25 million.
- Dana Wilkie
Interested in reading more?
